yasa.simulate_hypnogram¶
- yasa.simulate_hypnogram(tib=480, trans_probas=None, init_probas=None, seed=None, **kwargs)[source]¶
Simulate a hypnogram based on transition probabilities.
Current implentation is a naive Markov model. The initial stage of a hypnogram is generated using probabilites from
init_probasand then subsequent stages are generated from a Markov sequence based ontrans_probas.Important
The Markov simulation model is not meant to accurately portray sleep macroarchitecture and should only be used for testing or other unique purposes.
See also
New in version 0.6.3.
- Parameters
- tibint, float
Total duration of the hypnogram (i.e., time in bed), expressed in minutes. Returned hypnogram will be slightly shorter if
tibis not evenly divisible byfreq. Default is 480 minutes (= 8 hours).See also
- trans_probas
pandas.DataFrameor None Transition probability matrix where each cell is a transition probability between sleep stages of consecutive epochs.
trans_probasis a right stochastic matrix, i.e. each row sums to 1.If None (default), transition probabilities come from Metzner et al., 2021 [Metzner2021]. If
pandas.DataFrame, must have “from”-stages as indices and “to”-stages as columns. Indices and columns must follow YASA string hypnogram convention (e.g., WAKE, N1). Unscored/Artefact stages are not allowed.Note
Transition probability matrices should indicate the transition probability between epochs (i.e., probability of the next epoch) and not simply stage (i.e., probability of non-similar stage).
See also
Return value from
yasa.transition_matrix()- init_probas
pandas.Seriesor None Probabilites of each stage to initialize random walk. If None (default), initialize with “from”-WAKE row of
trans_probas. Ifpandas.Series, indices must be stages following YASA string hypnogram convention and identical to those oftrans_probas.- seedint or None
Random seed for generating Markov sequence. If an integer number is provided, the random hypnogram will be predictable. This argument is required if reproducible results are desired.
- **kwargsdict
Other arguments that are passed to
yasa.Hypnogram.Note
n_stages and freq must be consistent with a user-specificied trans_probas.
- Returns
- hyp
yasa.Hypnogram Hypnogram containing simulated sleep stages.
- hyp
Notes
Default transition probabilities are based on 30-second epochs and can be found in the
traMat_Epoch.npyfile of Supplementary Information for Metzner et al., 2021 [Metzner2021] (rounded values are viewable in Figure 5b). Please cite this work if these probabilites are used for publication.References
- Metzner2021(1,2)
Metzner, C., Schilling, A., Traxdorf, M., Schulze, H., & Krausse, P. (2021). Sleep as a random walk: a super-statistical analysis of EEG data across sleep stages. Communications Biology, 4. https://doi.org/10.1038/s42003-021-02912-6
Examples
>>> from yasa import simulate_hypnogram >>> hyp = simulate_hypnogram(tib=5, seed=1) >>> hyp <Hypnogram | 10 epochs x 30s (5.00 minutes), 5 stages> - Use `.hypno` to get the string values as a pandas.Series - Use `.as_int()` to get the integer values as a pandas.Series - Use `.plot_hypnogram()` to plot the hypnogram See the online documentation for more details.
>>> hyp.hypno Epoch 0 WAKE 1 N1 2 N1 3 N2 4 N2 5 N2 6 N2 7 N2 8 N2 9 N2 Name: Stage, dtype: object
>>> hyp = simulate_hypnogram(tib=5, n_stages=2, seed=1) >>> hyp.hypno Epoch 0 WAKE 1 SLEEP 2 SLEEP 3 SLEEP 4 SLEEP 5 SLEEP 6 SLEEP 7 SLEEP 8 SLEEP 9 SLEEP Name: Stage, dtype: object
Add some Unscored epochs.
>>> hyp = simulate_hypnogram(tib=5, n_stages=2, seed=1) >>> hyp.hypno.iloc[-2:] = "UNS" >>> hyp.hypno Epoch 0 WAKE 1 SLEEP 2 SLEEP 3 SLEEP 4 SLEEP 5 SLEEP 6 SLEEP 7 SLEEP 8 UNS 9 UNS Name: Stage, dtype: object
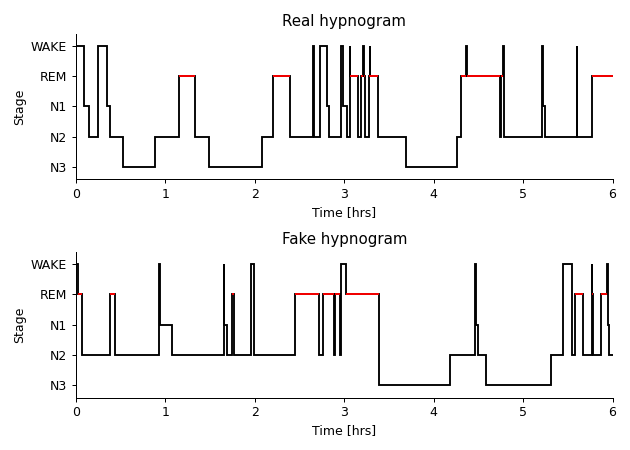
Base the data off a real subject’s transition matrix.
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from yasa import Hypnogram, hypno_int_to_str >>> url = ( >>> "https://github.com/raphaelvallat/yasa/raw/master/" >>> "notebooks/data_full_6hrs_100Hz_hypno_30s.txt" >>> ) >>> values_str = hypno_int_to_str(np.loadtxt(url)) >>> real_hyp = Hypnogram(values_str) >>> fake_hyp = real_hyp.simulate_similar(seed=2) >>> fig, (ax1, ax2) = plt.subplots(nrows=2, figsize=(7, 5)) >>> real_hyp.plot_hypnogram(ax=ax1).set_title("Real hypnogram") >>> fake_hyp.plot_hypnogram(ax=ax2).set_title("Fake hypnogram") >>> plt.tight_layout()