CONN Toolbox
Plot denoised BOLD timeseries of 2 ROIs
February 2017
A simple script to plot the BOLD denoised time-series of two specified ROIs from CONN Toolbox ROI.mat file
Define analysis info
- Info.wdir: path to your CONN/results/preprocessing folder which contains the denoised time-series of all the ROIs (see this post for a exhaustive review of CONN output files.
- Info.session: if you have several session (i.e. pre / post resting-state scans), enter the number of the desired session or put 0 for a temporal concatenation of all sessions.
- Info.nsub: number of subjects
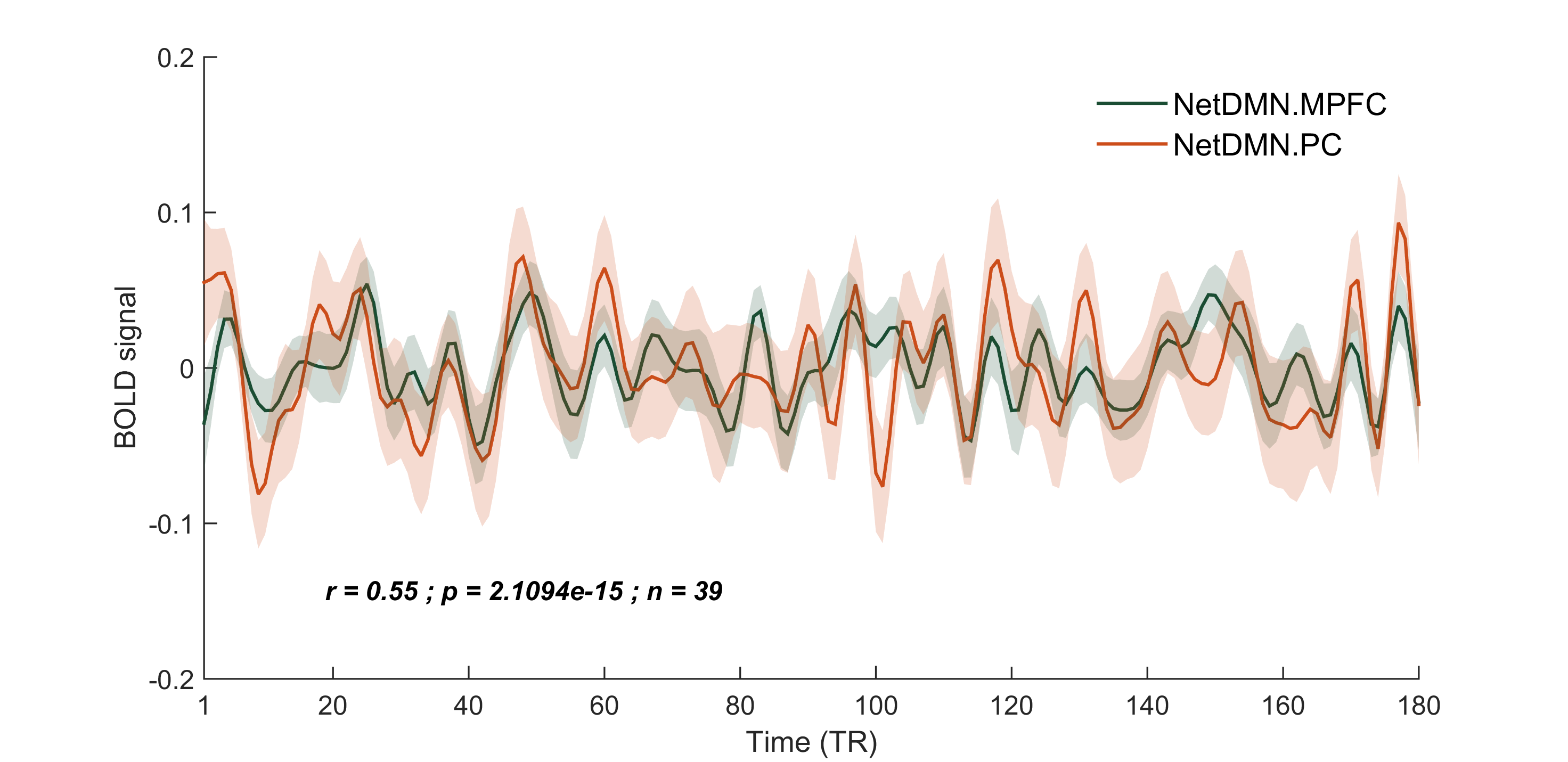
- ROI.ROIX_name: name of the ROIs you want to plot, as they appear in CONN Toolbox. In this example I am using two classic nodes of the default mode network, i.e. the medial prefrontal cortex and precuneous.
Info = [];
Info.wdir = 'C:/PhD/fMRI/CONN/conn_39s_preproc/results/preprocessing/';
Info.session = 1; % 0 for all sessions, 1=session1, 2=session2, etc...
Info.nsub = 39; % Total number of subjects
% ROI is the main structure containing ROI names, data
ROI = [];
ROI.ROI1_data = [];
ROI.ROI2_data = [];
% Select pair of ROIs
ROI.ROI1_name = 'NetDMN.MPFC';
ROI.ROI2_name = 'NetDMN.PC';
% Define outpath and outfilename
Info.outdir = pwd;
Info.outfile = [ Info.outdir '/TS_' ROI.ROI1_name '_' ROI.ROI2_name '_RUN'
num2str(Info.session) '.png' ];
Import data
Once this is done, the next step is to load the ROI mat file of each subject and find the two time-series of interest
for i=1:Info.nsub
% Loading .MAT file
matfile = [ 'ROI_Subject0', num2str(i, '%02i') ,'_Condition000.mat' ];
%fprintf('\nLoading:\t %s', matfile);
load([Info.wdir matfile]);
ROI.names = names;
ROI.dsess = data_sessions;
% Find index of selected ROIs
ROI.ROI1_idx = find(strcmp(ROI.ROI1_name, names));
ROI.ROI2_idx = find(strcmp(ROI.ROI2_name, names));
% Extract BOLD data
ROI.ROI1_data = [ ROI.ROI1_data , cell2mat(data(ROI.ROI1_idx)) ];
ROI.ROI2_data = [ ROI.ROI2_data , cell2mat(data(ROI.ROI2_idx)) ];
% Select sessions
if Info.session == 0 ;
ROI.cond = find(ROI.dsess);
else
ROI.cond = find(ROI.dsess == Info.session);
end
end
Compute means, sem and correlations
Then compute the mean timeseries with its corresponding standard error of the mean as well as the correlation coefficient between the two mean time-series.
% Compute mean, std and sem
ROI.ROI1_mean = mean(ROI.ROI1_data, 2);
ROI.ROI1_std = std(ROI.ROI1_data, 0, 2);
ROI.ROI1_sem = ROI.ROI1_std / sqrt(Info.nsub);
ROI.ROI2_mean = mean(ROI.ROI2_data, 2);
ROI.ROI2_std = std(ROI.ROI2_data, 0, 2);
ROI.ROI2_sem = ROI.ROI2_std / sqrt(Info.nsub);
% Correlation between mean timeseries
[rho_mean, pval_mean] = corr(ROI.ROI1_mean(ROI.cond), ROI.ROI2_mean(ROI.cond));
By default CONN toolbox applies a Fisher transform to all the R correlation values in order to homogenize the variance. If you prefer to compute the Z fisher-transformed value of your correlation coefficient, simply use the atanh(r) function of Matlab.
Plot
Mean time-series
% Create plot variables
ROI.x = [ 1:length(ROI.cond) ]';
ROI.ROI1_Y = ROI.ROI1_mean(ROI.cond,:);
ROI.ROI2_Y = ROI.ROI2_mean(ROI.cond,:);
ROI.ROI1_dy = ROI.ROI1_sem(ROI.cond, :);
ROI.ROI2_dy = ROI.ROI2_sem(ROI.cond, :);
set(0,'defaultfigurecolor',[ 1 1 1 ])
set(0,'DefaultAxesFontSize', 10)
fig = figure;
set(gcf,'Units','inches', 'Position',[0 0 6 3])
line_color = [ 0.1 0.3 0.2 ; 0.8 0.3 0.1 ];
set(gca, 'ColorOrder', line_color, 'NextPlot', 'replacechildren');
% Plot average ROI BOLD signal
plot(ROI.x, ROI.ROI1_Y, ROI.x, ROI.ROI2_Y, 'LineWidth', 1.5)
hold on
legend(ROI.ROI1_name, ROI.ROI2_name, 'Location', 'northeast')
legend('boxoff')
Semi-transparent continuous error bar
if Info.nsub > 1
fill([ROI.x;flipud(ROI.x)],[ROI.ROI1_Y-ROI.ROI1_dy;flipud(ROI.ROI1_Y+ROI.ROI1_dy)],
line_color(1,:),'linestyle','none', 'FaceAlpha', .2);
fill([ROI.x;flipud(ROI.x)],[ROI.ROI2_Y-ROI.ROI2_dy;flipud(ROI.ROI2_Y+ROI.ROI2_dy)],
line_color(2,:),'linestyle','none', 'FaceAlpha', .2);
end
Annotate the correlation value
dim = [.2 .2 .3 .1];
str = [ 'r = ', num2str(round(rho_mean, 2), '%.2f'), ' ; p = ',
num2str(pval_mean), ' ; n = ', num2str(Info.nsub) ];
annotation('textbox',dim,'String',str,'FitBoxToText','on', 'EdgeColor',
'none', 'FontWeight', 'bold', 'FontAngle', 'italic');
Set axis properties
y_lim = [ -0.2 0.2 ];
ylim(y_lim);
xlim([0 length(ROI.cond)]);
%set(gca, 'XColor', 'w'); % Mask x-axis
ylabel('BOLD signal');
xlabel('Time (TR)');
set(gca, 'XTick', [ 0:20:length(ROI.cond) ]);
grid off
box off
% Savefig 600 dpi
fig.PaperPositionMode = 'auto';
print(Info.outfile,'-dpng','-r600')
clearvars -except ROI Info
And that's it!
